The Single Cell Immunophenotyping Core (SCIC) Has Moved!
Our website has moved to a new address: https://voices.uchicago.edu/scic/
Please update your bookmarks and visit us at our new location for the latest information and resources.
Thank you!
Who we are:
The Single Cell Immunophenotyping Core (SCIC), also referred to as the Self-Service Genomic Core, is a user-operated, full-service facility that provides researchers with access to advanced instrumentation for genomic studies, with a specialized focus on single-cell immunophenotyping and genotyping of immune cells.
Our services include single-cell next-generation sequencing (NGS) platforms and quality control (QC) technologies, supporting high-quality data generation for genomic studies. All instruments are located in KCBD 10220E and 10220C and are accessible to the Chicago Area Research Community.
User Benefits:
- Unrestricted Access
Enjoy 24/7 access to instrumentation, including weekends and holidays. Once trained, users gain full autonomy to operate equipment independently. - Rapid Data Delivery
Streamline your workflow with same-day results for critical outputs (e.g., sequencing data), ensuring minimal disruption to your research timelines. - Skill Development
Acquire hands-on expertise in next-generation sequencing (NGS) and advanced genomic technologies, building competencies that strengthen your professional profile and career trajectory.
Time efficiency is our key differentiator. During high-demand periods—such as summer and academic breaks—our on-demand, walk-in model prioritizes expedited processing, making SCIC the optimal solution for projects requiring fast turnaround times without compromising data quality.
Core Technologies and Resources:
- Next-Generation Sequencing (NGS)
- Illumina Platforms: NextSeq 2000, and MiSeq sequencers.
- Nanopore Platforms: PromethION 2 Solo (P2 Solo) and MinION Mk1C and Mk1B.
- Single-Cell Library Preparation
- 10X Genomics Chromium X: Enables all 10X Genomics' high-resolution single-cell sequencing applications.
- Quality Control (QC) Devices:
- Agilent TapeStation 4200: Validates DNA/RNA preparations and library integrity. Reagent and disposable requirements for this instrument will be specified in upcoming protocols.
- BioRad CFX384 qPCR system: Supports high-throughput, real-time qPCR in 384-well plate format.
-
Biochemical Analysis
Vet Axcel Chemistry Analyzer: A compact, high-throughput clinical chemistry system designed for rapid and reliable analysis of biochemical parameters in serum and plasma. It is particularly useful in translational research and preclinical animal studies for metabolic, liver, kidney, and toxicology profiling.
-
Single-Cell and Gene Signature Database Integration App
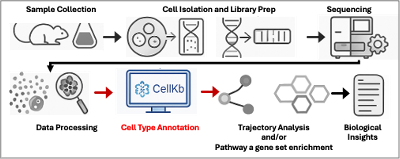
Cell Kb: A curated knowledge base of cell-type specific marker gene sets derived from published single-cell datasets. It supports cell type annotation, pathway insights, and reference-based interpretation of single-cell RNA-seq data, helping researchers contextualize their results more effectively.
- Expert Technical Support
- Experienced Personnel: Our dedicated team provides immediate assistance for troubleshooting experimental challenges, optimizing study design, and facilitating vendor communications.
- Peer Collaboration: We connect users with experienced peers (“master users”) conducting similar experiments to foster knowledge sharing and ensure technical success.
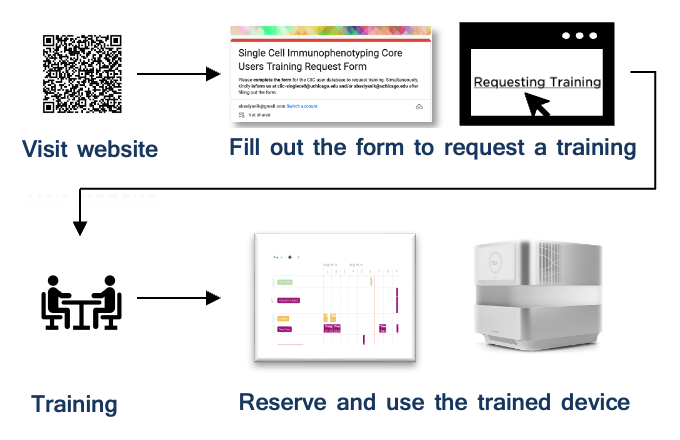
Getting Started: Device Access Workflow
- Initiate Access Request: New users should contact the SCIC team via email (ciic-singlecell@uchicago.edu) or directly reach out to the Core Scientist or Core Director (see below) to express interest.
- Submit Registration Form: Complete the required user registration form and notify the core team upon submission to initiate the onboarding process.
- Schedule Training: The SCIC team will coordinate a training session tailored to your selected instrument(s) and experimental needs.
- Complete Training: Participate in hands-on training with core personnel to gain proficiency in instrument operation, protocols, and safety guidelines.
- Activate BookitLab Access: Upon successful training, the SCIC team will grant you 24/7 scheduling privileges by registering your account in the BookitLab platform.
- Begin Independent Use: Reserve and operate instruments autonomously via BookitLab. Core personnel remain available for troubleshooting or advanced guidance.
Updates:
1. CellKb: Introducing a New Service for Core Users
We are excited to announce full enterprise access to CellKb, the world’s largest manually curated cell type marker database. CellKb Enterprise Service enables rapid, automated cell type annotation, prediction, and biomarker discovery at scale. Designed for cutting-edge transcriptomics and single-cell analysis, CellKb combines a robust knowledgebase, proprietary ranking algorithms, and a user-friendly interface to help researchers accelerate discoveries with accuracy and confidence.
With this new service, core users gain unlimited analyses, comprehensive cell type matching, full feature access, and premium support, empowering you to take your research further with reliable, harmonized cell type signatures and advanced annotation tools. Contact us at ciic-singlecell@uchicago.edu to get started with your enterprise account today. More info
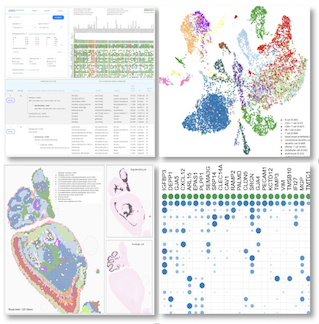
"CellKb generates accurate cluster annotations for single-cell data, removing the need for manual curation. It draws from thousands of curated signatures across studies with high-quality datasets, allows filtering by source organ and disease state, and returns fine-grained cell type annotations along with two parent-level classifications. This makes cluster annotation more reliable and broadly scoped than manually compiling reference datasets for marker comparison or dataset projection.", Maggie Clevenger, Bioinformatician
2. Vet Axcel Chemistry Analyzer Now Available at Our Core!
Precision Blood Chemistry for Marine Animal Research
The Vet Axcel Chemistry Analyzer is now optimized for marine animal studies, delivering rapid, accurate blood chemistry analysis to support research and conservation. With automated multi-parameter testing (including ALT, AMYL, BUN, and more), it reduces manual errors and streamlines workflows—ideal for high-throughput labs.
Trusted by Researchers:
“I’ve used the Vet Axcel® for serum analysis for years. It’s fast, reliable, and automates multiple parameters at once, cutting handling errors common in manual methods.”, Tatsuki Ueda, Post Doc Fellow
“As a Ph.D. candidate, time is critical. The Vet Axcel lets me analyze multiple analytes across samples with minimal pipetting. Affordable and high-quality data—fast.”, Madison Plaster, Ph.D. Candidate
"The blood chemistry analyzer is a high throughput, reliable method to produce comprehensive toxicity and metabolic profiling on blood and urine samples. In contrast to other methods like ELISA or individual assays, this method ensures high quality analysis of all potential contributors to systemic dysfunction in only a day.”, Taryn Beckman, UChicago PhD alumna

Announcement: N/A
Our core partnered with 10x Genomics to offer a hybrid Lunch & Learn event on April 27, 2022. This was an exciting opportunity to learn about 10x Genomics' current and upcoming single cell technologies, how to process samples at the Core, and see how single cell resolution can add depth to research questions. The speakers were Egon Ranghini (email, LinkedIn) from 10X Genomics, Michael Andrade from UChicago Medicine (email, LinkedIn), and our the Core Manager, Ha-Na Shim. Location: Eckhardt building, ERC 301B.
Please see the attached presentation and video link. PassCode: +B%$1eL2
A Scientific Seminar: Unifying Workflows: From Sample to Data
When: Wednesday, July 13, 2022
What time: 11:00 AM CT – 1:00 PM CT
Where: 1103, KCBD
Topic:
Single-cell analysis is a powerful technique to characterize complex tissue types, identify rare cell populations, uncover regulatory relationships between genes, and track cell trajectories. From sample handling to cell dissociation, there are a number of technical considerations that influence cell viability and data quality of single-cell genomics, single-cell epigenomics, and single-cell gene expression experiments.
Join us for this seminar, which covers critical sample prep decisions and technical considerations for single-cell assay design and optimization.
Agenda:
- 11:00am - 11:45am: General Sample Preparation Overview and Core Introductions
- 11:45am - 12:30pm: Breakout sessions 1-3
- 12:30pm - 1:00pm: Social and Group lunch
Breakout Sessions: Attendees split into 3 cohorts and will visit each Breakout session
• Organoids/Pathology (Le Shen and Chris Weber-Core)
• FACS (David Leclerc-Core and Miltenyi)
• Single Cell (Ha-Na Shim, Fatih Abasiyanik, Pieter Faber-Cores, and 10x Genomics)
The flyer and its power point presentation
Single Cell Immunophenotyping Core Seminar Series: Working with Sequencing Data: From Flow Cell to Single-Cell
Speaker: Jonathan MATTHEWS | Tay Lab, SCIC User; MEng, Electrical Engineering and Computer Science (BioEECS), MIT, 2016; BS, Electrical Eng. & Computer Sc., MIT, 2015
When: Wednesday, May 17, 2023
What time: 12:00 PM CT – 1:00 PM CT
Where: Zoom
Topics:
Join us for an informative seminar on the efficient handling of sequencing data in the RCC's flagship HPC clusters, Midway2 and Midway3, housed in the University of Chicago's data center. Our community benefits from state-of-the-art computing infrastructure that provides ample computing power and fast internet speed for enabling discovery and innovation.
The seminar will cover the following topics:
- Monitoring sequencing runs in BaseSpace using examples of good and bad quality check scores.
- Transferring BaseSpace Sequence Hub data, including Binary Base Call (BCL) and FASTQ files, to the RCC through Midway2 or Midway3 using the command line interface (CLI).
- Requeuing BCL files using the BaseSpace BCL Convert App.
- Uploading sequencing data from the RCC to the 10X Genomics Platform for processing using the Cell Ranger App.
Join us for this informative seminar to learn how to efficiently handle sequencing data in order to generate meaningful knowledge for scientific purposes.
Presentation File
Single Cell Immunophenotyping Core Seminar Series: Nanopore Applications: Next Generation Sequencing with NanoPore
Speaker: Dr. Filip Boskovic | Szostak Lab, SCIC User; HHMI Postdoctoral Associate and EMBO Postdoctoral Fellow, UChicago, HHMI; PhD, University of Cambridge, 2022

When: Tuesday, September 19, 2023
What time: 12:00 PM CT – 1:00 PM CT
Where: Zoom
Topics:
Join us for an informative seminar on the efficient handling of sequencing data in the RCC's flagship HPC clusters, Midway2 and Midway3, housed in the University of Chicago's data center. Our community benefits from state-of-the-art computing infrastructure that provides ample computing power and fast internet speed for enabling discovery and innovation.
The seminar covers the following topics:
- An Expedition through Nanopore Sequencing Advancements
- A Comparative Dive into Sequencing Technologies
- Masterful Tips and Tricks for Successful Nanopore Experiments
- Guided Tour of the General Sequencing Pipeline
- Unveiling Enhanced Data Analysis Techniques
- Accessible Links and Resources for Aspiring Nanopore Adventurers.
- Q&A (15 min.)
Join Us for an Informative Webinar on Nanopore Sequencing Technology. Discover the potential of Nanopore sequencing and its applications in modern genomics research.
Whether you're new to technology or looking to enhance your skills, this webinar is for you.
10x Genomics summarized their spatial omics tools and noval upgrades such as Visium-HD. You will learn how Chromium Single Cell, Visium Spatial, and Xenium In Situ platforms from 10x Genomics can help you push the boundaries of your research. Uncover molecular insights, dissect cell-type differences, investigate the adaptive immune system, detect novel subtypes and biomarkers, and map the epigenetic landscape cell by cell. Enabling deeper insight into cancer, immunology, neuroscience, and developmental biology, 10x Genomics gives researchers the ability to see biology in new ways.
- Speakers: Egon Ranghini (email, LinkedIn) from 10X Genomics, Michael Andrade from UChicago Medicine (email, LinkedIn), and Leilani Marty Sentos.
- Organizers: Fatih Abasiyanik (abasiyanik@uchicago.edu) and Heather Eckart (heather.edkart@10cgenomics.com)
- Location: Eckhardt building, ERC 301B.
- Date & Time: Wednesday, January 10, 2024; 14:00-15:30
Video Link and Passcode: r^5i*KQ
This is a special hybrid seminar organized jointly by the Single Cell Immunophenotyping Core and Illumina!
The seminar will focus on introducing the XLEAP-SBS™ Chemistry on the NextSeq™ 2000 and the NovaSeq™ X Plus instruments.
Seminar Details:
- Date: 15th May, 2024
- Time: 10:00-11:30
- Location: William Eckhardt Research Center, Room 201B
- Topic: Introducing XLEAP-SBS™ Chemistry on the NextSeq™ 2000 and the NovaSeq™ X Plus
- Speakers: Logan Silber (Sequence Sales Specialist) and Chevelle Blackburne (FAS)

Event Objectives:
- Understand the features and benefits of XLEAP-SBS™ Chemistry
- Explore the enhanced performance of the NextSeq™ 2000 and NovaSeq™ X Plus instruments
- Gain insights into designing experiments to leverage the latest advancements in sequencing technology
Seminar Details:
Title: Spatial Protein Phenotyping with Phenocycler-Fusion Technology
Date: May 29, 2024, 11:00 AM -12:00 PM; Location: ERC 201 B
Speaker Lineup:
-
Avik Mukherjee, PhD, Sr. Technical Applications Scientist, Akoya Biosciences
-
Surya Pandey, PhD, Northwestern University Immunotherapy Assessment Core
Characterizing the complexities of the tissue microenvironment is critical in understanding the molecular and cellular mechanisms driving disease and therapeutic response. Protein spatial phenotyping enables the determination of which cells are present, their location, their biomarker patterns, and how they are organized and interact with each other. The PhenoCycler-Fusion™ (formerly CODEX®) technology provides a comprehensive solution for whole-slide, single-cell, spatially resolved, multiplexed immunofluorescence, allowing quantitation and spatial visualization of 80+ protein markers. This high-throughput format complements spatial transcriptomics data and can significantly advance your research.
Coffee Chat: Best Practices in Sample Preparation for Single Cell Sequencing
- Date & Time: Wednesday, November 13, 2024 | 9:30 AM – 11 AM CT
- Location: KCBD Auditorium, 1103, 900 E. 57th St., Chicago, IL 60637
Agenda Highlights:
- 9:30 - 9:45 AM: Coffee and treats
- 9:45 - 11:00 AM: Seminar featuring speakers from 10x Genomics and Miltenyi Biotec, along with service overviews from the CAT Facility and the Single Cell Immunophenotyping Core
- Post-Seminar Workshops and Tours:
- Workshop Session 1: 11:15 - 12:15 PM
- Workshop Session 2: 12:30 - 1:30 PM
Each workshop included a 20-minute tour of the CAT Facility, followed by time for Q&A.
The teams from 10x Genomics and Miltenyi Biotec were available to discuss specific details and address your project-specific questions.
Please find the flyer. This workshop was designed to enhance data quality for single cell sequencing workflows, and we believe it will be beneficial to many in the research community.
Recordings:
Part 1: Best Practices in Sample Preparation for Single Cell Sequencing-MILTENYI
Part 2: Best Practices in Sample Preparation for Single Cell Sequencing-10X Genomics
Illumina's AI algorithms are transforming our approach to drug discovery and precision medicine. In this session, we will explore how DRAGEN™ leverages machine learning to enhance the accuracy of variant calling. We will also examine the data showcasing the impact of the PrimateAI-3D algorithm on our understanding of variants of unknown significance (VUS), and how explainable AI in Emedgene™ software streamlines germline tertiary analysis workflows.
- Date & Time: May 15, 2025 | 10:00 AM – 11 AM CT
- Location: KCBD Auditorium, 1103
- Speaker: Danieal Saul, Information Specialist, Illumina, Inc.
Recordings:
- Date & Time: July 16, 2025 | 10:00 AM – 11 AM CT
-
Speaker: Ashwini Patil, CEO Combinatics l PhD Bioinformatics
Highlights from the Session
Live Demo: Dr. Patil performed a real-time walkthrough of CellKb’s interface, processing and annotating actual 10X Genomics and Visium spatial data during the session.
From Raw to Result: The session demonstrated how to filter, cluster, and annotate scRNA-seq data using Seurat and Scanpy, showing how CellKb can integrate seamlessly into modern workflows.
Tool Comparison: Attendees gained insights into how CellKb outperforms traditional annotation methods, offering higher confidence and literature-backed results.
Exclusive Access: Participants were given early access to CellKb’s annotation services and priority onboarding for their upcoming single-cell projects.
🎥 Watch the Recording
Missed the live session or want to revisit it?
The full recording is now available on YouTube: ▶️ Watch the Webinar Recording
It was an insightful talk on generating ultra-rich data and meaningful insights with Nanopore Sequencing.
It covers :
- Target enrichment from amplicons to adaptive sampling
- Direct DNA/RNA sequencing with built-in methylation detection (no bisulfite needed)
- Full-length transcript sequencing for isoform and fusion detection at the bulk or single-cell level
- Date & Time: July 30, 2025 | 10:00 AM – 11 AM CT
- Location: ERC, 201B
- Speaker: Madeline Hartley, Sr. Field Applications Scientist at Oxford Nanopore Technologies





